Our latest publication on the Schistoscope was highlighted in the Journal of Medical Imaging.
Automated medical imaging framework revolutionizes schistosomiasis diagnosis
New framework powers rapid and accurate detection of Schistosoma haematobium eggs.
Schistosomiasis, a parasitic disease affecting millions worldwide, poses a significant public health and economic burden, particularly in impoverished regions. To combat this disease and achieve World Health Organization (WHO) targets for control and elimination, accurate and accessible diagnostic tools are essential. Currently, microscopy is the standard for diagnosing schistosomiasis, but it is time-consuming, operator-dependent, and requires specialized expertise, making it challenging for resource-limited areas.
To address these challenges, researchers developed the Schistoscope, an innovative optical tool equipped with an autofocusing and automated slide scanning system. This device captures microscopy images of urine samples, enabling efficient detection of Schistosoma haematobium eggs, a common cause of urogenital schistosomiasis. In a study published in the Journal of Medical Imaging, the researchers aimed to create a robust dataset and develop a two-stage diagnostic framework using deep learning to accurately identify and count S. haematobium (SH) eggs in field settings.
First, the researchers created an SH dataset consisting of 12,051 images of urine samples collected in a rural area in central Nigeria and captured using the Schistoscope device. They manually annotated the images, marking the eggs and differentiating them from artifacts such as crystals, glass debris, air bubbles, and fibers, which can hinder accurate diagnosis.
The proposed two-stage diagnostic framework consists of a DeepLabv3 with a MobilenetV3 backbone deep convolutional neural network, trained using transfer learning on the SH dataset. In the first stage, the framework performs semantic segmentation to identify candidate SH eggs in the captured images. The second stage refines the segmentation by fitting overlapping ellipses, effectively separating boundaries of clustered eggs, leading to more accurate egg counts.
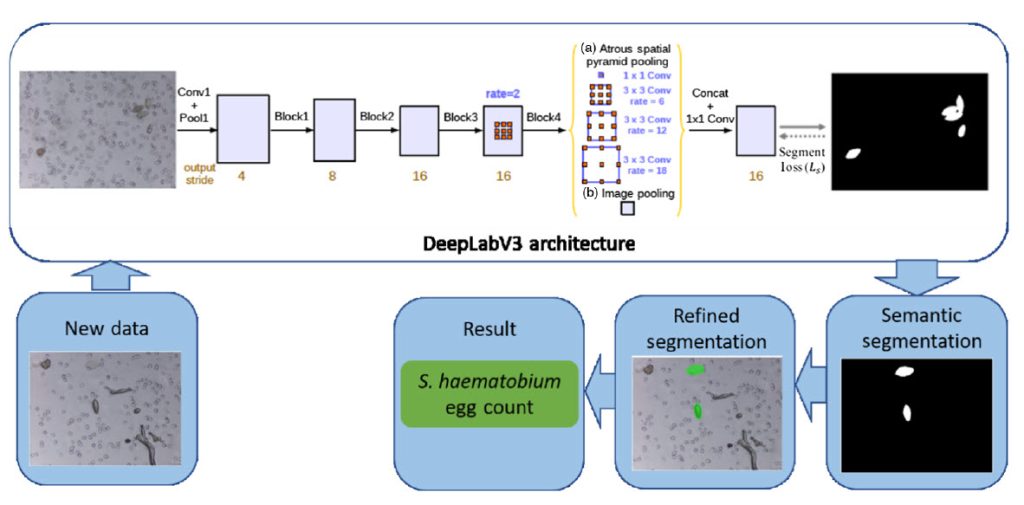
To demonstrate the field applicability of the proposed framework, the researchers implemented it on an edge AI system (Raspberry Pi + Coral USB accelerator) and tested it on 65 clinical urine samples obtained in a field setting in Nigeria. The results showed high sensitivity, specificity, and precision (percentages: 93.75, 93.94, and 93.75, respectively), with the automated egg count closely correlated to the manual count by an expert microscopist.
This SH dataset serves as a valuable resource for training and evaluating the diagnostic framework, providing a diverse set of images with varying degrees of difficulty due to artifacts.
Professor Jan Carel Diehl, of Delft University of Technology’s Department of Sustainable Design Engineering and corresponding author for the study, remarks, “By automating the egg detection process, the Schistoscope and the proposed diagnostic framework offer a promising solution for the rapid and accurate diagnosis of urogenital schistosomiasis, particularly in low-resource settings. Future studies will further validate the framework’s performance and compare it with other diagnostic methods, such as schistosome circulating antigen detection and DNA-based assays, to establish its role in schistosomiasis monitoring and control.”
Overall, this work represents a significant step towards improving diagnostics and combating schistosomiasis, a disease that disproportionately affects vulnerable populations in endemic regions.
Read the Gold Open Access article by P. Oyibo et al., “Two-stage automated diagnosis framework for urogenital schistosomiasis in microscopy images from low-resource settings,” J. Med. Imag. 10(4) 044004 (2023), doi 10.1117/1.JMI.10.4.044005